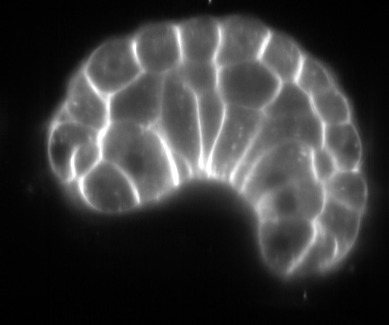Section: New Results
3D Modeling of developing organisms
Participants : Gaël Michelin, Grégoire Malandain, Léo Guignard [Virtual Plants] , Christophe Godin [Virtual Plants] .
This work is made in collaboration with Patrick Lemaire (CRBM).
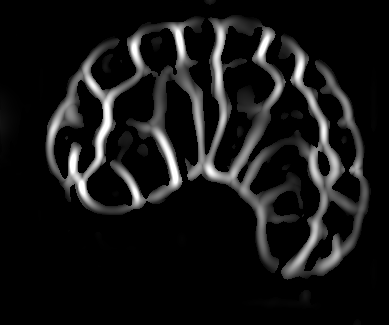
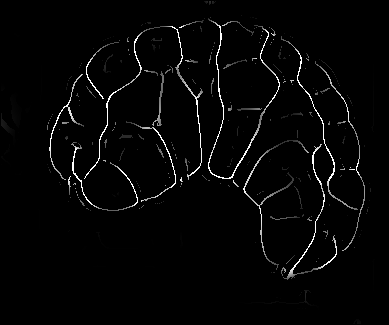
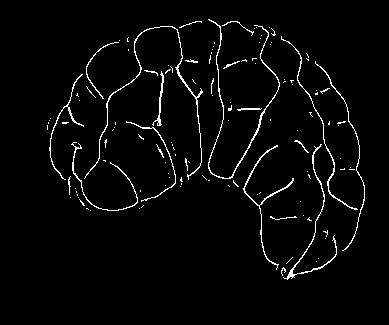
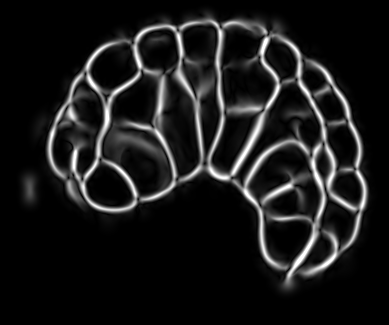
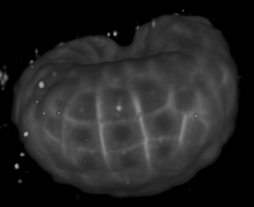
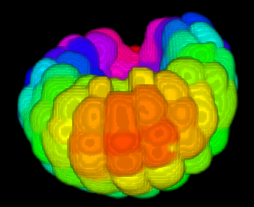
Image-based studies of developing organs or embryos produce a huge quantity of data. To handle such high-throughput experimental protocols, automated computer-assisted methods are highly desirable. We aim at designing an efficient cell segmentation method from microscopic images. Similary to another work [32] , the proposed approach is twofold: first, cell membranes are enhanced or extracted by the means of structure-based filters, and then perceptual grouping (i.e. tensor voting) allows to correct for segmentation gaps (see figure 6 ). We assessed different structure-based filters as well as different perceptual grouping strategies to identify the most efficient combination, in term of result quality and computational cost [13] .
|